MLE Subcommands
The main entry point for using the mle-toolbox is via its subcommands. Similar to how you would call git <subcmd>, you can use the different functionalities of the toolbox via mle <subcmd>. In the following we will go through all of the available subcommands and disuss their usage and optional inputs.
mle run - Experiment Execution 🚀
An experiment can be launched from the command line via:
mle run <experiment_config>.yaml
The <experiment_config>.yaml file provides all experiment settings such as the experiment type and resources required per individual job. Read more about how to configure it here. You can add several command line options, which can come in handy when debugging or if you want to launch multiple experiments sequentially:
--no_welcome: Don't print welcome messages at experiment launch.--no_protocol: Do not record experiment in the PickleDB protocol database.--resource_to_run <resource>: Run the experiment on the specified resource.--purpose: String purpose to store in the protocol database.
The --purpose option is especially useful if you would like execute multiple experiments sequentially. In this case you would like to run a bash script and not have to specify the purpose at launch time. E.g.
mle run exp_1.yaml --purpose Experiment 1 Grid
mle run exp_2.yaml --purpose Experiment 2 Grid
By default the jobs will run on your local machine. If you specify a different <resource> you will be asked whether you would like to sync your code with a remote directory. Afterwards, the experiment will continue to be executed on that resource.
mle monitor - Monitor Resources 🖥️
mle monitor provides a simple wrapper around the mle-monitor dashboard. It will load the protocol database using the configuration settings provided in ~/mle_config.toml and the resource you are currently working on. Afterwards, the dashboard will be updated in real time and synchronized with most recent protocol from the GCS bucket every other minute.
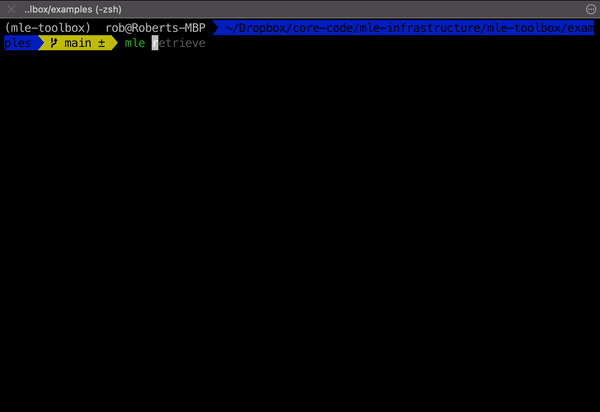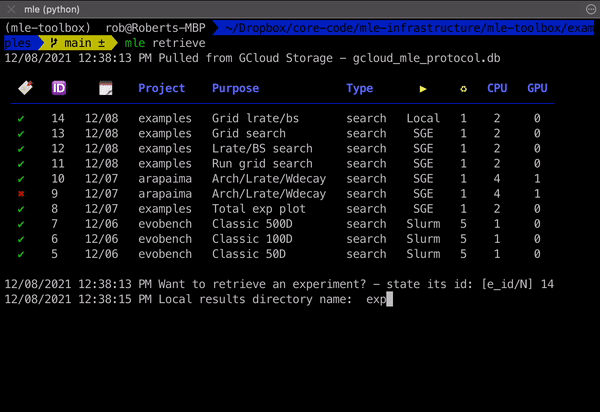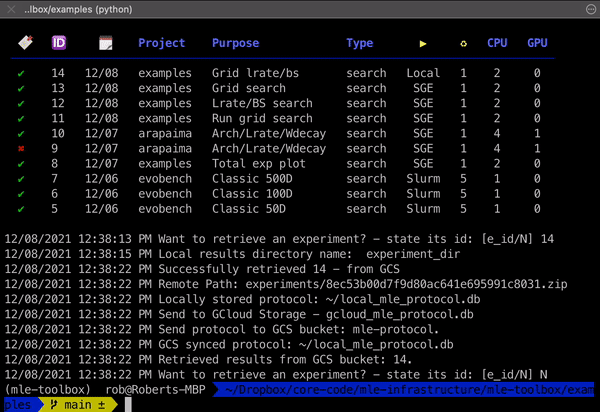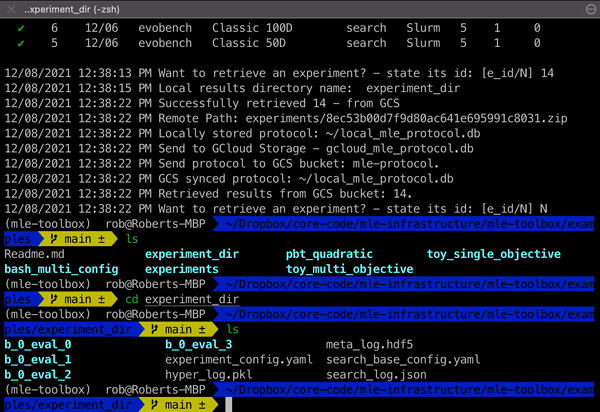
mle retrieve - Retrieve Results 📥
Once your experiment has finished, you can retrieve its results (logging directory) using the mle retrieve subcommand. This will fetch the corresponding remote/GCS storage path from the protocol database and then copy the results over.
mle report - Report Results 💌
mle report will create a set of report files (.html, .md & .pdf) corresponding to your experiment. The files will contain the experiment configuration and figures generated from the experiment logs. Alternatively, you can also set report_generation to true in the meta_job_args of your experiment .yaml file. In this case the report will be automatically created once your experiment is completed. If you are using the slack bot notification feature the report will be send to your slack account.
mle init - Setup Toolbox Configuration ⏳
When you first start to setup the toolbox, you can run mle init in order to create a mle_config.toml file that is stored in your root directory. It automatically launch your favorite editor so that you can modify your configuration settings.
mle sync - Download GCS-Backed Experiments 🔄
If you have run multiple experiments over night and you would like to retrieve all experiments from your GCS bucket which were not synced previously, you can simply run mle sync. The toolbox will retrieve all zipped experiment directories and unpack them for you.
mle project - Initialize New Template Project 🗂
I noticed that most of my projects follow the same file structure. It is depicted in the mle-project repository. If you run mle project, the toolbox will clone the repository, delete all git files and rename the directory, so that you can directly get started coding up your project with MLE-Infrastructure support.
mle protocol - List State of Protocol 📝
You can get a snapshot of your most recent experiments which were protocolled by running mle_protocol. This can be useful if you don't want to run the dashboard, but simply want to get a quick overview.